Welcome to PySB: Systems biology modeling in Python
PySB is a framework for building mathematical models of biochemical systems as Python programs. PySB abstracts the complex process of creating equations describing interactions among multiple proteins or other biomolecules into a simple and intuitive domain specific programming language (see example below), which is internally translated into BioNetGen or Kappa rules and from there into systems of equations. PySB makes it straightforward to divide models into modules and to call libraries of reusable elements (macros) that encode standard biochemical actions. These features promote model transparency, reuse and accuracy. PySB also interoperates with standard scientific Python libraries such as NumPy, SciPy and SymPy, enabling model simulation and analysis.
For further information, please see the following publication:
Lopez, C. F., Muhlich, J. L., Bachman, J. A. & Sorger, P. K. Programming biological models in Python using PySB. Mol Syst Biol 9, (2013). doi:10.1038/msb.2013.1
Download
To get started with PySB, see the download page for instructions.
Documentation
All of the PySB technical documentation is available online, courtesy of Read the Docs. The pysb/examples directory in the source tree contains sample models and other scripts if you'd prefer to learn by example. One of these examples and its output is reproduced below.
For further information, or to contact the developers, see the support page.
Contributing
Development is centered around our GitHub project page. Bugs and other problems should be reported at the issue tracker, and we welcome pull requests for fixes and enhancements.
Example
Model code:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 | |
Output:
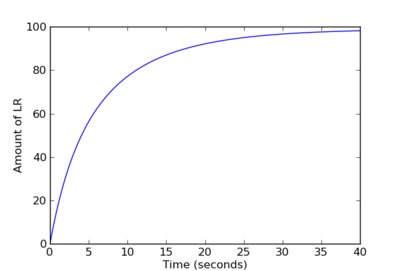
The above source code is available as hello_pysb.py in the directory of example models and scripts included with the PySB distribution. Note that this simple model does not demonstrate macros or any other higher-order features of PySB modeling. For an advanced model, see EARM 2.0.
Authors
PySB is primarily developed by members of the Lopez Lab at Vanderbilt University and the Sorger Lab at Harvard Medical School:
Vanderbilt
- Alex L. R. Lubbock (alex.lubbock@vanderbilt.edu)
- Leonard A. Harris (l.harris@vanderbilt.edu)
- James C. Pino (james.c.pino@vanderbilt.edu)
- Carlos F. Lopez (c.lopez@vanderbilt.edu)
Harvard
- Jeremy L. Muhlich (jmuhlich@bitflood.org)
- John A. Bachman (bachmanjohn@gmail.com)
Funding
PySB has been supported by the National Institutes of Health under award numbers P50GM68762 (PK Sorger), P01CA139980 (PK Sorger) and K22CA151918 (CF Lopez). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This material is based upon work supported by the National Science Foundation under Grant Number 1411482 (CF Lopez). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
